Introduction
Every summer, South Korea faces many problems owing to algal blooms (
NIER 2020;
Ahn et al. 2021a), which cause the death of aquatic life and negatively affect water supply management (
Schmidt et al. 2013;
Pavagadhi and Balasubramanian 2013;
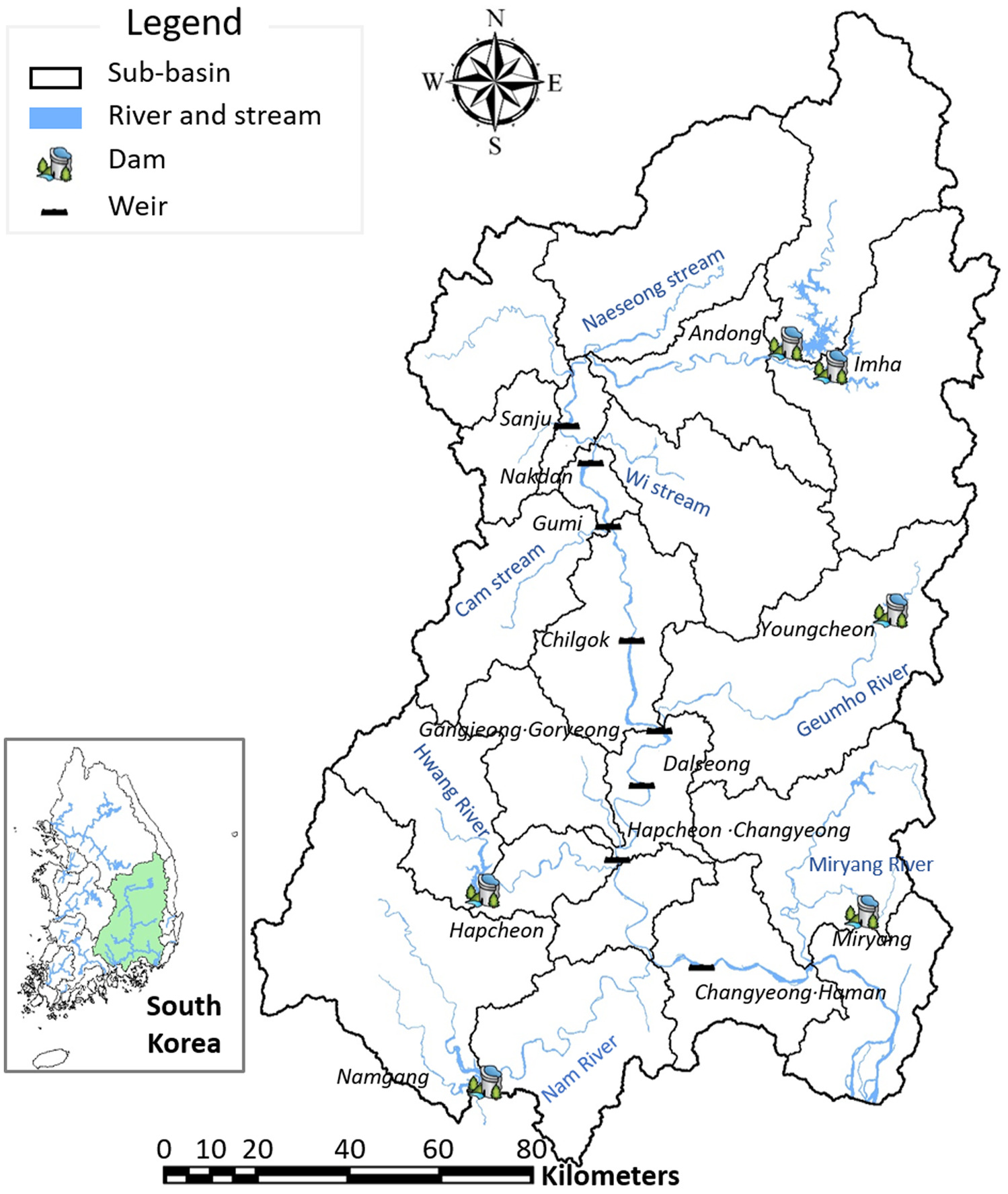
Preece et al. 2017). In particular, because the Nakdong River has been transformed artificially into a lake due to the Four Major Rivers Project, the occurrence of cyanoHABs has become even more severe. Algal blooms are a global environmental issue typically occurring when cyanobacteria proliferate (
Huisman et al. 2004). In South Korea, algal blooms caused by cyanobacteria also are a serious social problem (
Ahn et al. 2021c). Therefore, predicting cyanobacteria cell counts is crucial for managing cyanobacterial harmful algal blooms (cyanoHABs) (
Pyo et al. 2020).
In South Korea, water quality and cyanobacteria cell counts are measured weekly for early response and management of cyanoHABs. These data are used to predict cyanobacteria cell counts. The physical Environmental Fluid Dynamics Code (EFDC)–National Institute of Environment Research (NIER) model predicts cyanobacteria cell counts. The EFDC-NIER model has been improved by the National Institute of Environmental Research to better suit South Korean river environments and has undergone extensive improvements for enhanced prediction accuracy (
Ahn et al. 2021b). The EFDC-NIER model is utilized for two main purposes: short-term forecasting for operating the harmful algae alert system; and predicting variations in cyanobacterial occurrences based on various scenarios involving meteorological, hydrological, water quality, and hydraulic factors. It is crucial to predict the occurrence of harmful algal blooms (HABs) in the near future quickly and accurately. By doing so, sufficient time can be secured to implement proactive measures and strategies to effectively mitigate the impacts of HABs on river ecosystems and minimize potential risks to water supply safety in surrounding communities. The ability to make timely and precise short-term forecasts allows for better planning and preparation, thus enhancing the overall management and response to HAB events.
Operating a harmful algae alert system using a physics-based model can present several challenges. One of the major issues is the computational complexity and time required to execute the model. The process of simulating the physical interactions and environmental factors is resource-intensive and time-consuming. Running simulations to predict algal blooms and their dynamics involves complex calculations that require substantial computational power and data processing. Input data, model structure, and parameters considerably influence cyanobacteria cell count predictions using physics-based models (
McAvoy et al. 2003;
Zhang et al. 2015;
Jiang et al. 2018;
Su et al. 2018). Physics-based models can yield accurate results through well-validated mechanisms and processes. However, the complexity of these mechanisms and processes can make model construction difficult and result in significantly longer execution times. As a result, obtaining real-time or near-real-time predictions may be difficult, hindering the system’s ability to provide timely warnings and effective responses to potential HAB events. This limitation calls for the exploration of alternative modeling approaches or techniques that can strike a balance between accuracy and computational efficiency in order to improve the operational capabilities of the harmful algae alert system.
Our main research objective was to leverage the strengths of artificial intelligence (AI)-based deep learning prediction models to complement the weaknesses of the physics-based models. For this purpose, we applied artificial intelligence technology to predict harmful algal blooms and compared the results with those obtained from traditional physics-based models. Through discussions and analysis, we present the limitations of AI technology and propose potential solutions to overcome them, ultimately enhancing the accuracy and efficiency of algal bloom predictions.
AI-based deep learning models exhibit strengths in handling large-scale data processing and providing rapid predictions (
Lek and Park 2008). By learning patterns and relationships from past data, they enable quicker response times and more-efficient short-term forecasting (
Miotto et al. 2018). Therefore, data-driven models can be an alternative to physics-based models because they require only data distribution to predict cyanobacteria cell counts (
KEI 2020). Research on cyanoHABs is relatively scarce, and it is still a very challenging field. However, several studies have been conducted recently. Neural-network-based AI technology mainly was used in those studies (
Velo-Suarez and Gutierrez-Estrada 2007;
Coad et al. 2014;
Zhang et al. 2015). ANN techniques have been used widely in algae prediction, including for predicting cyanobacteria cell counts (
Maier et al. 1998;
Xiao et al. 2017;
Yim et al. 2019;
Chen et al. 2020). ANNs have evolved in various ways depending on the purpose of prediction. In particular, long short-term memory (LSTM) solves the gradient vanishing problem and improves performance when predicting long sequences (
Kim et al. 2021). Cyanobacteria data and various weather and water-quality variables affecting cyanobacteria have a time-series distribution. LSTM is particularly effective for time-series analysis and is used widely in predicting algae (
Lee and Lee 2018;
Shin et al. 2019;
Hill et al. 2020;
Liang et al. 2020;
Zheng et al. 2021). Recently, convolutional neural networks (CNNs) have been used to predict the temporal and spatial distribution of cyanobacteria (
Pyo et al. 2020). Reports indicate that CNNs have advantages in understanding the features of cyanobacteria data. Accordingly, recent studies show that models combining a CNN and LSTM yield the best results compared with other neural-network-based models in cyanobacteria prediction (
Naghdi et al. 2020;
Cao et al. 2022).
In previous studies using CNN-LSTM to predict cyanobacteria, the focus was on utilizing the advantages of two-dimensional (2D) CNNs for image processing to extract features from cyanoHABs regions along with meteorological data, and then using LSTM for prediction. In other words, the studies aimed to forecast the spatial variation of algal blooms in lakes. Our main objective was to predict the cyanobacteria directly as a numerical value, specifically in terms of cell counts. This approach represents a significant advancement, because it allows for more-precise and quantitative predictions of cyanobacteria occurrences, which can be crucial for effective management and mitigation strategies. Comparing the results of the AI-based prediction model with those of the conventional physics-based model in regions with relatively scarce water quality and algal bloom data, such as Korea, is a critical aspect of our research. The significance of our research lies in evaluating the applicability of artificial intelligence in the fields of water quality and algal blooms, which have not been explored extensively compared with the existing evaluations focused on floods and droughts. Although the utilization of AI has been assessed in flood and drought aspects by comparison with physics-based models, our study provides a fresh perspective by focusing on water quality and algal bloom prediction.
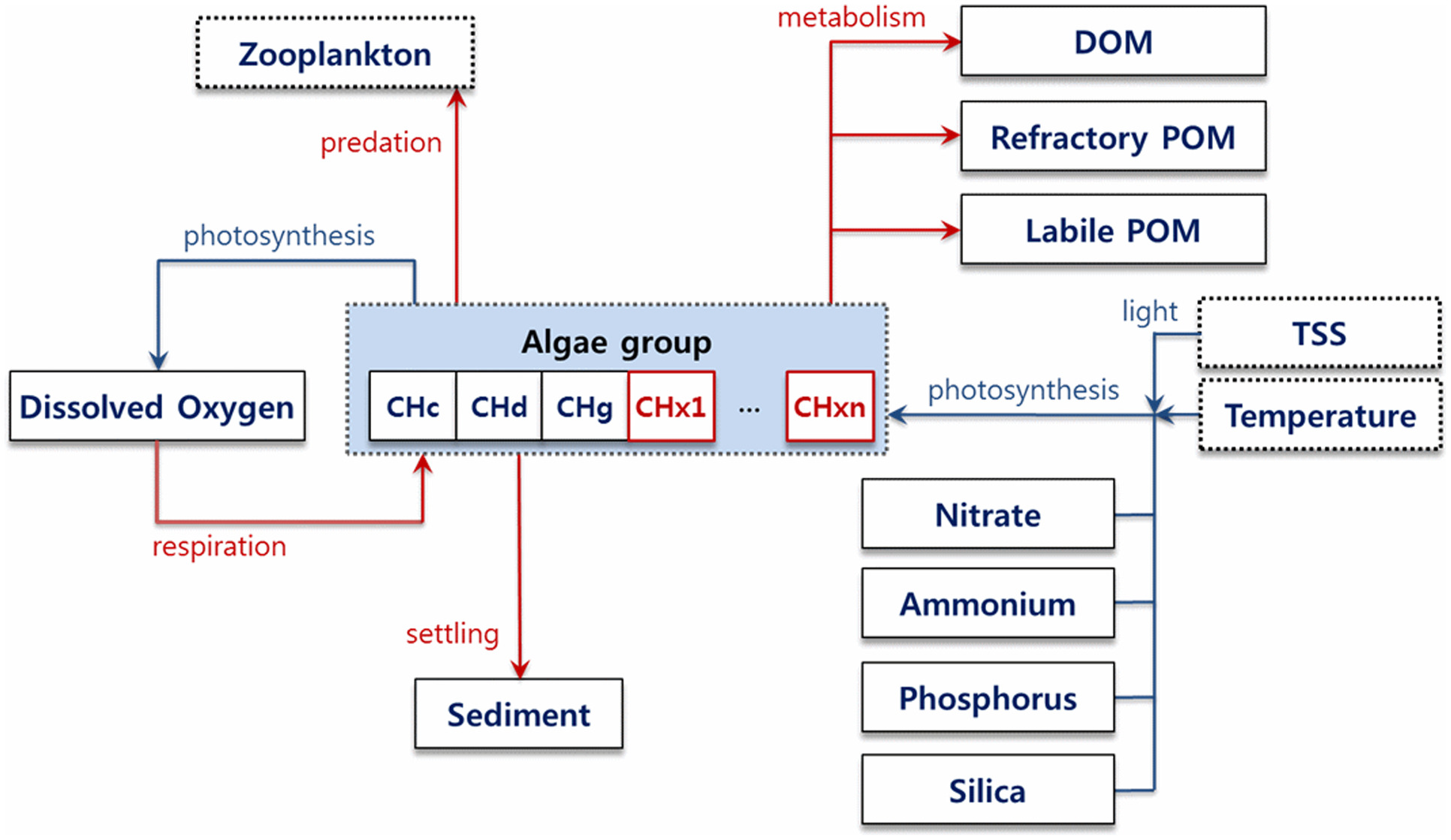
In this study, we selected the CNN-LSTM model, a combination of a CNN and LSTM, as the AI-based deep learning model to compare with the physics-based model. When choosing the optimal model, it is crucial to consider the characteristics of the data that are to be predicted. Cyanobacteria cell counts exhibit a unique feature in which the observed data can vary drastically from 0 to as high as several hundred thousand
, unlike other variables. Recently, several studies have suggested that combining LSTM with a CNN can enhance accuracy in predicting time-series data with rapid and drastic changes (
Hwang and Shin 2020). A CNN helps identify meaningful patterns and correlations in the data (
Han et al. 2019), and LSTM can effectively capture temporal dependencies, thus enhancing the overall predictive performance (
Wan et al. 2020;
Xia et al. 2020). This combination of a CNN and LSTM allows the model to benefit from the complementary nature of these two architectures, leading to improved accuracy and more-robust predictions in time-series forecasting tasks, especially when dealing with data that exhibit rapid and drastic changes.
The primary contributions of this study are as follows:
1.
In the existing research on predicting harmful algal blooms, the focus has been on predicting the spatial distribution of harmful algae. In contrast, our study improved the model to predict cyanobacteria cell counts directly.
2.
The developed AI-based deep learning model was compared with the conventional physics-based model that traditionally is used in the field of cyanoHAB prediction. We advanced the evaluation process beyond comparing the predictions with the observed data by directly comparing the results of the AI-based model with those of the established physics-based model. This comprehensive comparison allowed us to thoroughly assess the applicability and limitations of the AI model in the context of predicting cyanobacteria cell counts.
3.
By evaluating the performance of AI-based models in these specific domains, our research contributes to a broader understanding of the potential and limitations of artificial intelligence in cyanobacterial forecasting and management.
Discussion
Limitation of the CNN-LSTM Model
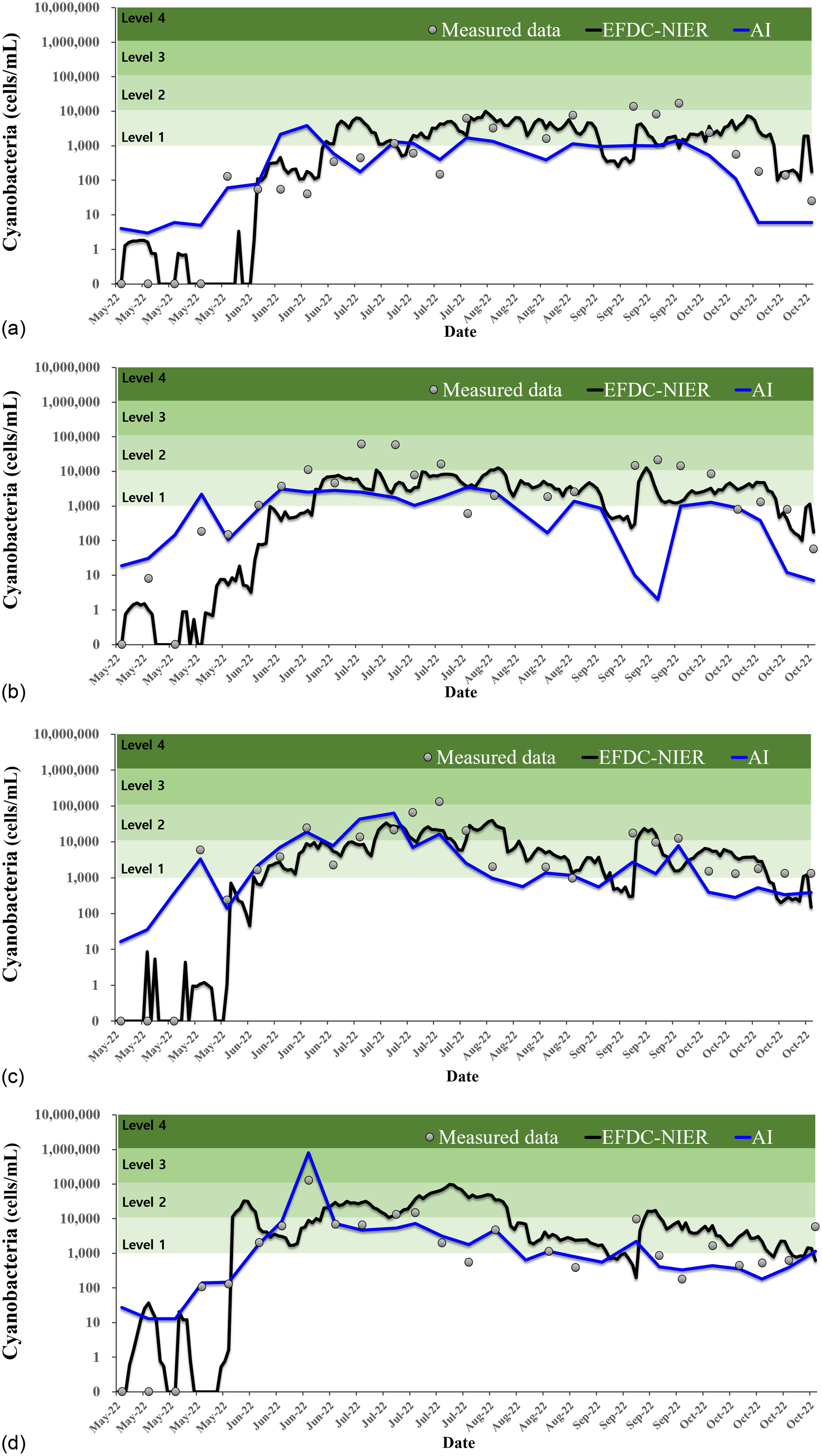
The comparison of the two models showed that the CNN-LSTM model outperformed the EFDC-NIER model overall. However, the CNN-LSTM prediction had a major limitation. The predictive power was significantly low for Level 4 (). If the HAB alert is at Level 3 () or Level 4, cyanoHABs have occurred and an alert has been issued. At Level 4, special management is required owing to the severe cyanoHAB levels. From this perspective, the CNN-LSTM model with lower predictive power in Level 4 than the existing EFDC-NIER model can be considered to be a significant limitation in terms of managing cyanoHABs.
The first cause of this limitation is the overfitting problem. To prevent this overfitting problem, a dropout rate of 0.2 was set in this study (
Shah et al. 2018). However, the amount of training data for Level 4 was very inadequate compared with that for the other levels. Consequently, although highly accurate training results were obtained, the accuracy in the prediction interval decreased because of overfitting.
The second cause is that the pattern of cyanobacteria occurrence in 2022 was different from that in the past. In 2022, a severe drought and heat wave occurred in the Nakdong River Basin. In particular, Gyeongsangbuk-do and Gyeongsangnam-do, where the Nakdong River is located, experienced the lowest number of rainy days in the last 10 years, according to the Korea Meteorological Administration’s climate information portal.
As a result, since 2018, the worst cyanoHABs occurred in 2022. In particular, for the first time since monitoring began, Gumi Weir in the upstream area of the Nakdong River had cyanobacteria cell counts of more than in 2022. This is a substantially different phenomenon from past patterns, in which a large amount of cyanobacteria was not observed in the upper reaches.
The CNN-LSTM is a technique for predicting the future by learning past time-series patterns. Therefore, the predictive power may be lower for future patterns that have not occurred in the past. Deep-learning models, such as the CNN-LSTM, are black box models in which AI autonomously extracts desired information without specifying complex multidimensional data by users (
Sengupta et al. 2020). Understanding the learning process of these deep-learning algorithms is difficult, making the identification of the causes that affect uncertainty challenging. By contrast, physics-based models, such as the EFDC-NIER model, can simulate HABs using well-defined equations and mathematical theories.
Applicability of AI-Based Deep Learning Models in the Prediction of Cyanobacteria Cell Counts
Although the CNN-LSTM model has limitations, it demonstrated excellent performance in predicting cyanobacteria cell counts. It achieved results that are comparable to or slightly better than those of the physics-based models, such as the EFDC-NIER model, while significantly reducing the analysis time. In general, cyanobacteria cell counts are predicted by collecting real-time measurement data in the morning and analyzing them; then forecasters make a prediction in the afternoon based on the results. However, the construction of input data for the EFDC-NIER model requires a considerable amount of time, and the analysis takes several hours or more. Moreover, human errors or incorrect model designs can lead to situations in which the results cannot be produced within the deadline. The CNN-LSTM model provides the advantage of having a prediction time of a few minutes. Therefore, even if an error is present in the input data configuration, it can be reanalyzed easily. In addition, CNN-LSTM models have the advantage of being analyzed easily even by nonexperts. The EFDC-NIER and other physics-based models typically rely heavily on the model’s structure and parameters (
Jiang et al. 2018;
Su et al. 2018). Therefore, building such models and adjusting the parameters require substantial expertise. In contrast, data-driven models, such as CNN-LSTM, can easily construct input data, making them accessible to the general public.
Proposing a Direction for the Research on AI-Based Deep Learning Models in the Prediction of Cyanobacteria Cell Counts
The recent trend of the various developments and advancements in AI algorithms has revealed that the application of deep-learning-based AI technology in the management of cyanoHABs is inevitable. This paper presents both the positive aspects and limitations of deep-learning-based AI technology. Measures that can be taken to overcome the limitations and apply deep-learning-based AI technology to cyanoHABs management are as follows.
Through comparison with the results of the physics-based model, the AI-based deep learning model demonstrated the capability to construct rapid models and predict cyanobacteria cell counts based on data statistics even in situations with limited prior knowledge. In situations in which there are sufficient data, the AI-based deep learning model has shown comparable or better results than the physics-based model, especially in cases with low cyanobacteria cell counts. However, in cases in which data are limited, the AI-based deep learning model demonstrated lower predictive performance for new data, because it may not capture the relationships between input and output variables sufficiently, particularly in cases with high cyanobacteria cell counts. The findings of this study provide evidence that there is a need for the complementary use of the physics-based model and the AI-based deep learning model.
AI-based deep learning models allow for rapid model construction and predictions based on data statistics even in situations with limited prior knowledge. However, they can be challenging to interpret physically, and their reliability may be dependent on the conditions included in the training data, exhibiting a tendency to rely heavily on the provided data (
von Stosch et al. 2014). Conversely, physics-based models, grounded in first principles, focus on learning interpretable causal relationships between input and output variables, aiming to understand the physical world. These models significantly advance the understanding of various phenomena in fields such as material balances, fluid dynamics, and reactor kinetics, thereby addressing the limitations of AI-based deep learning models mentioned earlier. Physics-based models offer the advantage of providing a deeper understanding of the physical world, and can complement the drawbacks associated with AI-based deep learning models (
Daw et al. 2021). Physics-based models also may face challenges in model construction due to insufficient prior knowledge or complex mechanisms (
von Stosch et al. 2014;
Jeong et al. 2023). The drawbacks associated with physics-based models can be complemented by AI-based deep learning models. Therefore, AI-based deep learning models and physics-based models possess distinct characteristics and have the potential to mutually complement each other’s limitations, because they offer unique strengths that can address the weaknesses of the other (
Aykol et al. 2021).
To overcome the limitations of AI-based deep learning models, as a general solution, collecting and constructing more input data for the model can be effective. This is not an easy task, considering the substantial budget and effort required to collect and observe data. However, by applying the concept of hybridization, combining the strengths of both AI-based deep learning models and physics-based models, it is possible to address these challenges and find solutions. A well-calibrated physics-based model can predict cyanobacteria cell counts accurately at Level 4 because of its internal mechanisms and various parameters, even with limited data. In addition, it can be used to analyze scenarios that did not exist in the past, such as dam and weir operations in rivers, virtual climate scenarios, and water intake in river. If the simulated results are utilized in a AI-based deep learning model, they can complement the shortage of input data and partially solve the problem of overfitting. This approach, which has been proposed widely in recent research, takes advantage of the clear boundaries between AI-based deep learning models and physics-based models (
Vo et al. 2022;
Jung et al. 2023). As a result, it allows for the application of existing methodologies specific to each model type. This hybrid method harnesses the unique strengths of both approaches, enabling a more versatile and effective solution that can accommodate different modeling requirements and enhance overall predictive capabilities (
von Stosch et al. 2014;
Aykol et al. 2021). The fusion of these models can lead to a more robust and comprehensive approach, offering improved predictive capabilities and a better understanding of complex systems while overcoming data limitations in certain scenarios.
A direct combination of the two models, not just a simple fusion concept, also can serve as an alternative to overcome limitations. This hybrid model, by integrating both models directly, has the potential to address the drawbacks effectively. Because examples of combining hydrological analysis with deep learning exist in the field of hydrology (
Maraun et al. 2017;
De Luca et al. 2020), combining the mechanisms of cyanobacterial growth, proliferation, and extinction with deep-learning-based AI algorithms seems possible. In this concept, the hybrid model can incorporate a loss function that reflects the physical laws present in the physics-based model. This enables the AI-based deep learning model’s predictions to be controlled, ensuring that they do not violate the physical laws (
Wang et al. 2019). One of the key advantages of this approach is that it allows for the interpretation of predictive results in a physically meaningful way, leveraging the benefits of both models.
However, when applying this hybrid concept, if the accuracy of the predictions from the physics-based model is not sufficiently ensured, using them as inputs for the AI-based deep learning model can lead to increased uncertainty in the results. Therefore, to ensure the accuracy and reliability of the results, further research is needed on the calibration of parameters in the physics-based model. Additionally, efficient deep learning algorithm structures need to be developed to incorporate the mechanisms and processes, such as the physical laws present in the physics-based model, into the algorithms of the AI-based deep learning model. Such efforts are essential to enhance the overall performance and effectiveness of the hybrid approach.